This article is designed to provide you with guidance, insights, and explanations for unexpected results you may have experienced with your bacterial transformation and gel electrophoresis kit. Select the lab you are having issues with to find an explanation and/or solution:
Bacterial Transformation
Select the issue you have experienced to learn possible causes.
Issue: No transformed bacteria grew on my selective (antibiotic-containing) plates.

Potential Causes
- Were the bacteria used to perform the transformation (i.e. starter plate) incubated for too long? Maximum 20 hours post-inoculation if incubating at 37°C and 40 hours post-inoculation if incubating at room temperature.
- Were single colonies selected for transformation? See from time 0:00–0:10 in the video below on how to select for single colonies. Overgrowth on starter plates makes selecting single colonies difficult. To prevent overgrowth, starter plates should be streaked for isolation using the correct technique.

- Was the plasmid DNA added to the +plasmid tubes? See time 0:35 in the video below.
- Is there growth on the control plate? If not, ask if the LB broth was added after transformation. If this is not done, the bacteria become stressed and are unlikely to grow.
- Was the heat shock procedure done correctly? Moving directly and quickly from ice to water bath for 90 seconds then directly and quickly back to ice is essential. Extended time between ice and water, such as walking across a room between the two, can have a negative impact. If not, then the plasmid DNA may not have entered the cells, as the heat shock step facilitates the entry of plasmid DNA into the cells.
Issue: I have transformed colonies on my selected plate, but they are not showing the color phenotype (GFP for pGreen; blue for pBLU; glow-in-the-dark for pVIB).
Potential Causes
- pGreen: pGreen plasmid contains a mutant version of GFP that turns bacteria yellow-green, even in normal light. If you expose the colonies to a UV light, they also fluoresce, but do not glow in the dark. Incubation temperature can influence expression of the GFP trait as well. If incubation temperatures are held above 37°C, GFP expression is not consistent.
- pBLU: Was the X-gal added to the LB Amp media? If not, the blue color will not be expressed, because the expression of the beta-galactosidase gene is controlled by an inducible promoter that is turned on only when X-gal is present.
- pVIB:
- Were the transformant organisms incubated at 30°C? If they were incubated at higher temperatures, this may have prevented folding of the proteins that cause fluoresce.
- Sometimes the luminescence of the bacteria can be difficult to see. For optimal viewing, take plates into a completely dark space and allow your eyes a few minutes to adjust for observation.
Issue: I observe small colonies around my transformed colonies that look different.

Potential Cause
- These are called satellite colonies and will sometimes appear. The antibiotic ampicillin works by inhibiting bacterial growth rather than killing bacteria, causing any bacteria that have been plated and not transformed to be present on the plate in small numbers. Transformed bacteria that express ampicillin resistance conferring protein will secrete this protein around their colonies, causing ampicillin in the media around them to break down. This creates regions on the plate where non-transformed bacteria can grow, which are the small satellite colonies you are observing.
Issue: An excessive number of bacteria grew on my +plasmid and -plasmid plates.
Potential Causes
- Was the media cooled to around 55°C before the antibiotic was added? If not, then the antibiotic is not functional and will not select for transformed colonies.
- Was the antibiotic added to the media? This is necessary for selection of transformants.
Issue: I have few transformant colonies on my +plasmid plate.
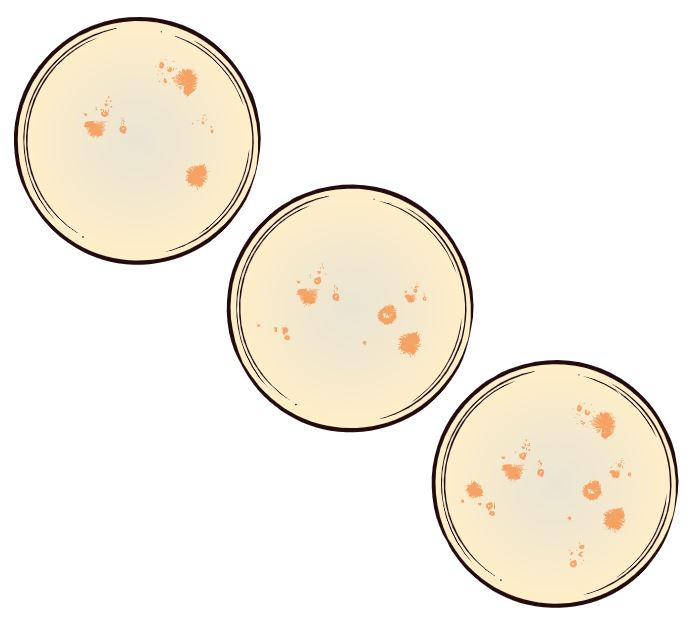
Potential Cause
- Did you follow the lab protocol for recovery time post-heat shock? In our experience, reduced recovery times decreased the number of obtained colonies. Other deviations from the lab protocol can impact this as well.
Gel Electrophoresis
Select the issue you have experienced to learn possible causes.
Bands are missing from a lane or lanes on my gel
Potential Causes:
Was the gel run for a longer time or at a higher voltage than specified by the protocol?If so, this can cause bands of DNA to run further down or even off of the gel, causing them to not be visible. Always check that your lab equipment and run time are aligned with recommended lab protocols and timing. Additionally, check that the dye front is monitored for the best results.


Did a well get punctured by the pipette when loading DNA? If a pipette is inserted too deeply when loading DNA into a well, it may puncture through a gel, causing DNA to be lost through the hole. You may be able to observe a pipette tip-sized hole in your wells if this has occurred. Best practice is to have your students practice pipetting before performing a gel. We recommend our practice pipetting stations kit.


Issue: Bands do not move far down the gel or appear to not separate (compressed).
Potential Cause:
Was the gel run for a shorter time or at a lower voltage than specified by the protocol? If so, this can cause bands to not move very far down a gel and can cause bands to not separate. Always check that your lab equipment and run time are aligned with recommended lab protocols and timing.


Issue: Bands appear faint and are difficult to see.
Potential causes:
Could a smaller volume of the sample than specified by the protocol been loaded? Running a gel with very small samples of DNA or dye will make bands difficult to visualize. Alternatively, a sample may have been poorly loaded, causing some of the sample to come out of the well, or an air bubble may have been inserted below a sample in the well, causing the sample to leave the well as the bubble floats up out of the well.


Was a smaller amount of restriction enzymes than called for used with your digest? If so, this can cause faint bands and/or larger bands due to the smaller-than-desired number of digests by restriction enzymes. Performing digest for a smaller or longer time than recommended can also cause this issue. It is always best to closely follow recommended digestion parameters.


Issue: Bands are smeared or streaky.
Potential causes
Was more DNA than required added to your digest? Loading a well with a large volume of DNA can cause smearing or streaks to appear behind bands. This could also be caused by running a gel at a higher-than-recommended voltage.
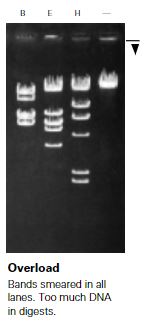

When removing the comb from your gels, did the wells appear damaged or not fully formed? Wavy or smeared bands can occur when a sample moves through a non-uniform well. This most often occurs when a comb is removed before a gel is completely set.


Was your gel made only with water or the incorrect concentration of buffer? If so, this difference in concentration between your gel and the buffer solution it is submerged in can cause smeared or streaky bands.


Issue: I see powder or small clumps of a substance on my gel.
Potential cause:
Using a different ratio of buffer powder to distilled water than specified by a protocol when preparing buffer solution can cause a precipitate (solid) to form in your solution, creating a snowy or flaky appearance within your gel. Be sure to check that no crystallization or precipitate has formed when your buffer solution is prepared before using it to prepare gels or to fill gel boxes.







2 Comments
This lab worked well, but it would be VERY helpful if specific directions were given for viewing results with the various plasmids. I was expecting to see bacteria that were glowing in the dark, and had to search this site to find that our pGREEN plasmid produces yellow green colonies that DO NOT glow in the dark, but rather show fluorescence under a UV light. The UV light was not provided in the kit, nor was it listed in additional materials needed but not provided.
Hello, and thank you for the feedback! It is customers like you who help keep our kits up to date and providing feedback to help improve them. Additionally, we do offer a plasmid that confers glowing in the dark to bacteria, pVIB (Item #211447), which is in our glow-in-the-dark transformation kits (item# 211088P, 211088) and hope you try this kit in the future to obtain the results you desire. UV lights are omitted from our kits to keep our kit costs low, because they are not needed to visually identify green colonies to confirm transformation, and to prevent teachers from receiving items they may already have. We appreciate your suggestion to add UV light to the “needed but not required” section of our website for these kits and will do so as an optional item