
Which Genetic Organism Is Best for Your Classroom?
Which Genetic Organism Is Best for Your Classroom? With so many genetic organisms to choose from, it can be difficult to find the organism that
Bioinformatics is a fusion of biology, computer science, and information technology focused on analyzing biological data and developing sophisticated software tools to draw conclusions and make comparisons using data from different sources. With this rapidly emerging technology, researchers are now able to learn more about how a change in protein structure alters protein function and can also easily compare entire genomes of different organisms.
Due to the large volumes of DNA data being generated each day in labs around the world, scientists have developed a specialized system of managing the information. NCBI, the National Center for Biotechnology Information is a U.S. government agency that serves as a clearinghouse for biological resources. The NCBI website includes more than 30 individual databases available to the public, providing powerful opportunities for your students to engage in bioinformatics in the classroom.
One of NCBI’s commonly used search tools is the Basic Local Alignment Search Tool (BLAST). BLAST searches can be used to find proteins, transcripts, primers, and gene expression profiles from within the NCBI database. This means that in the classroom, students using BLAST are able to:
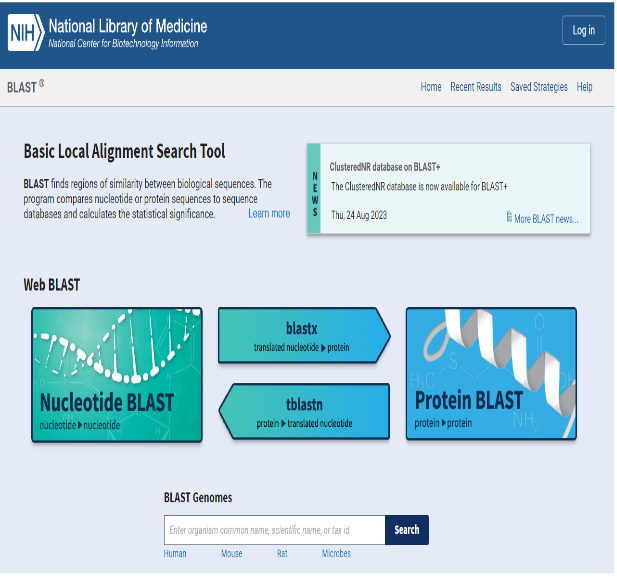
This article provides an introduction on how to use BLAST in the classroom in addition to outlining hands-on activities available from Carolina that include bioinformatic exercises using BLAST and DNA Subway, a bioinformatics platform from our partners at the DNALC at Cold Spring Harbor.
Note: Be aware, and make your students aware, that NCBI’s website is updated frequently and that some of the details and screens may have changed since this article was written.
Credit: Courtesy of the National Library of Medicine. Please see the NIH website granting permission for use of their images and works.

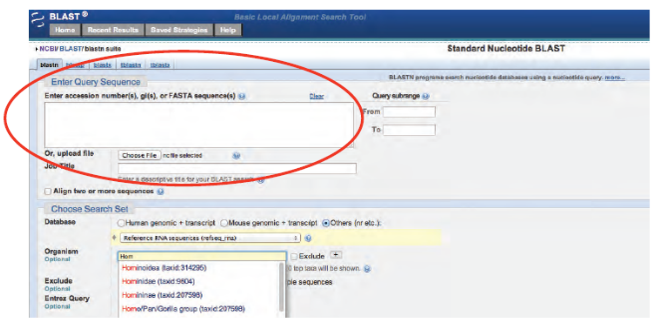
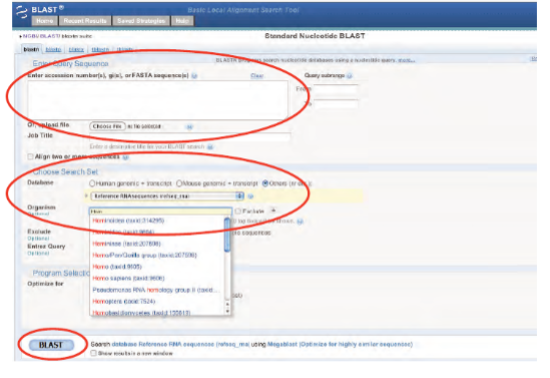
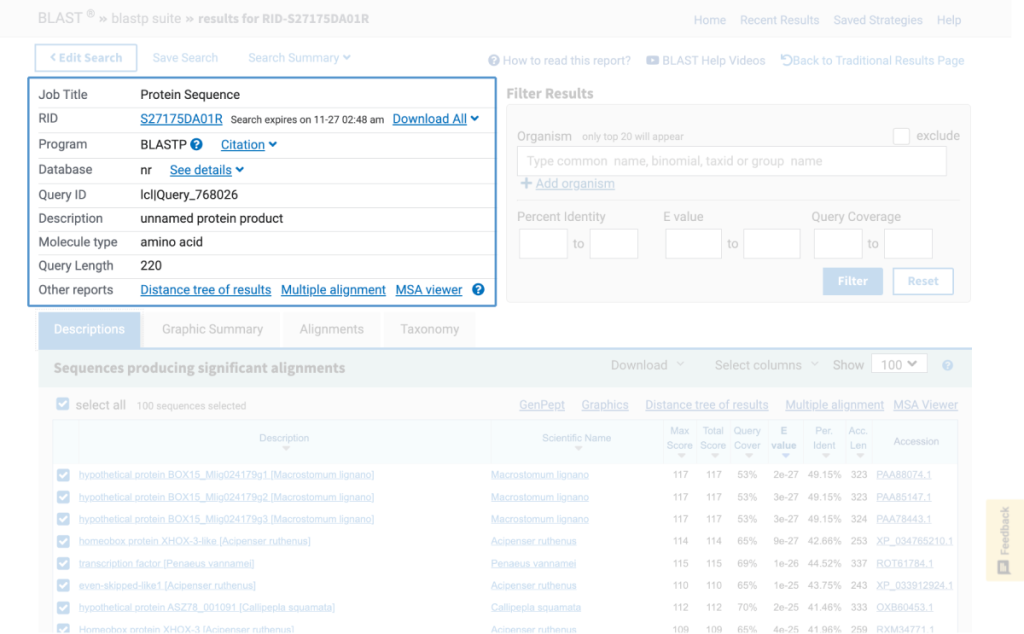
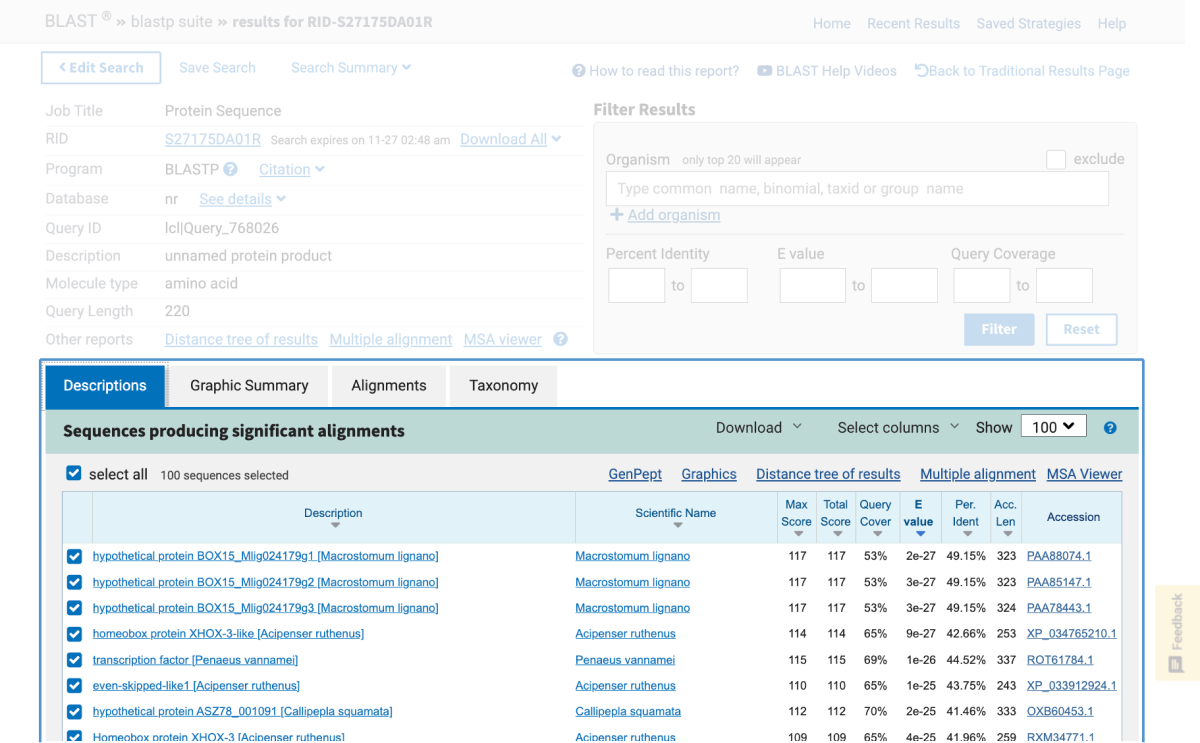
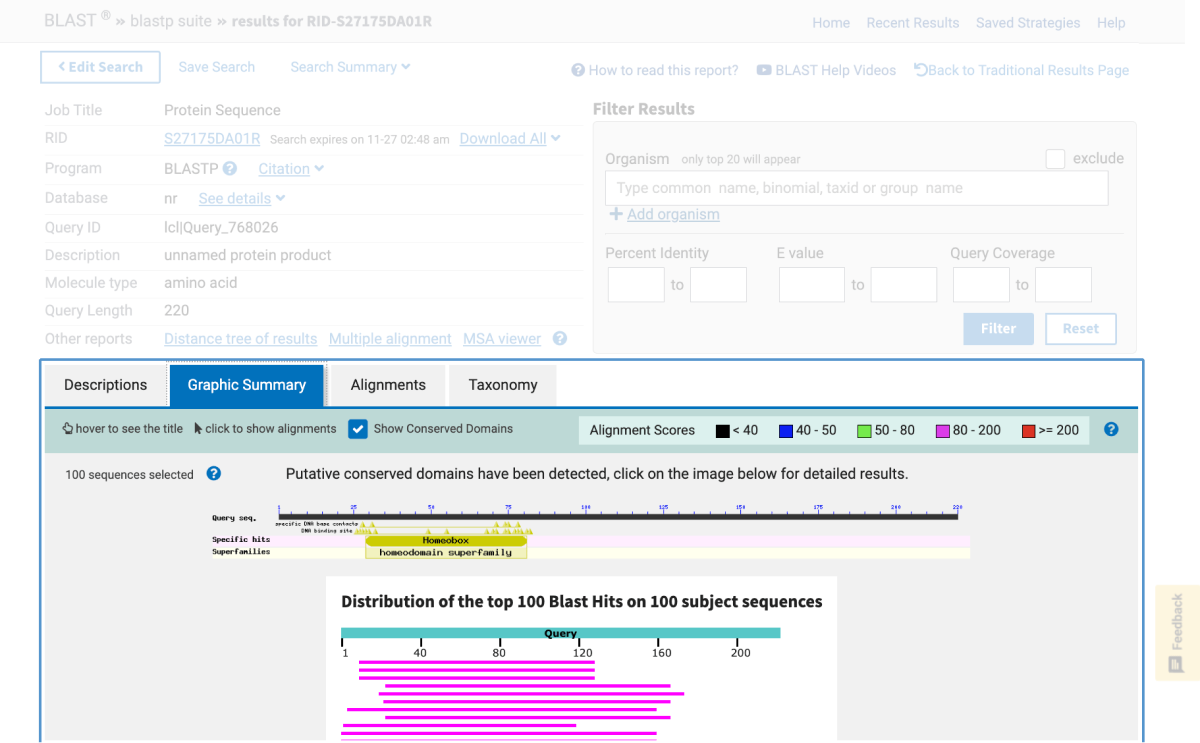
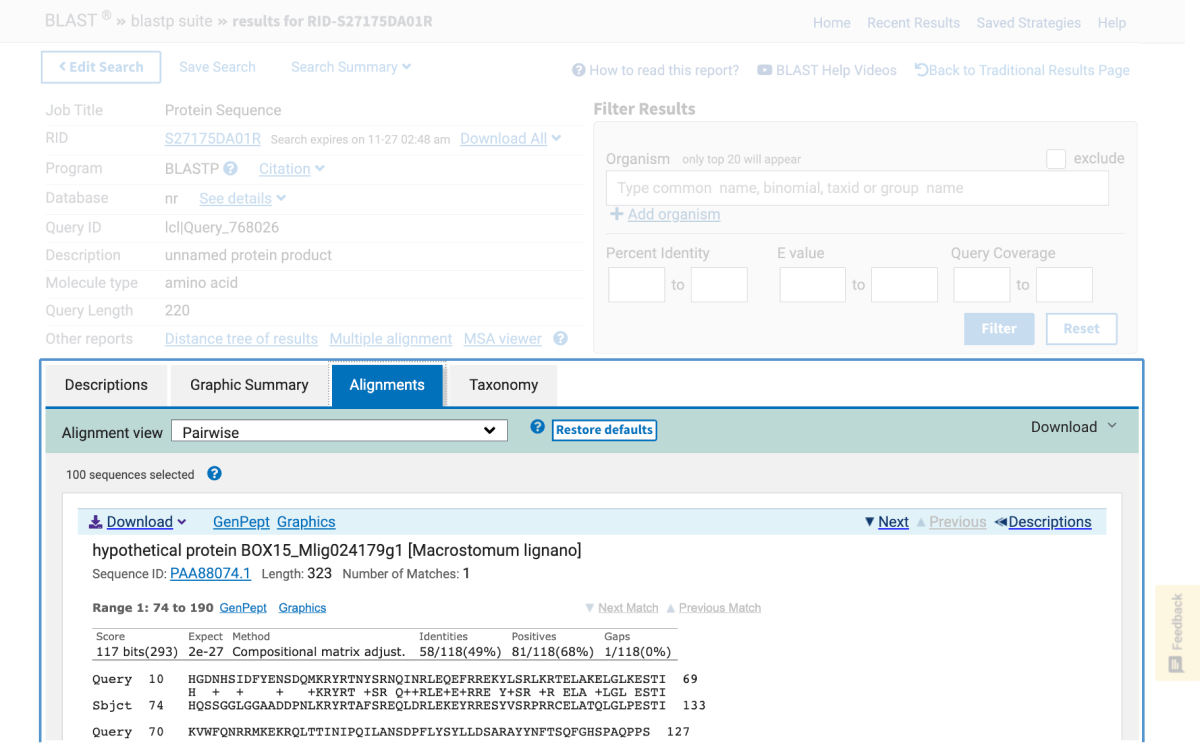
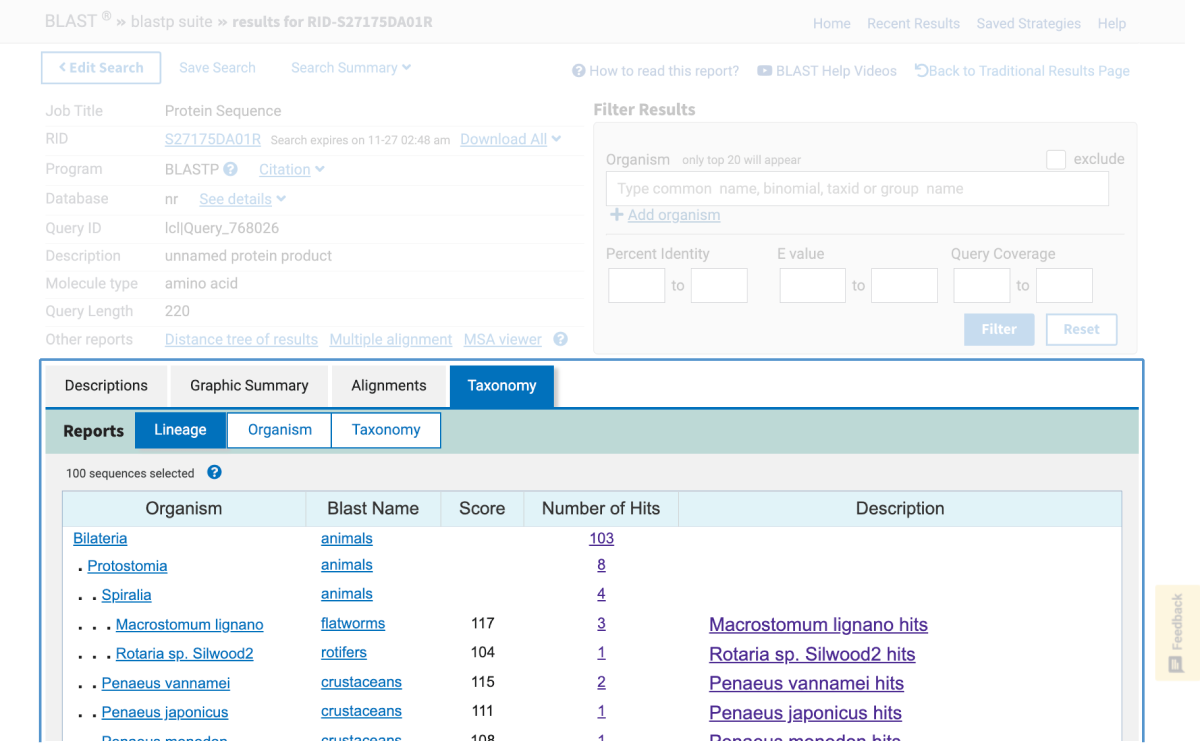
Note: Bioinformatics databases and analytics change frequently, sometimes without notice. This is why for each kit below, if needed, there is a revised, digital update for performing the activity.
Bioinformatics Activities:
Location of the PV92 Alu Insert
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211230A.pdf
This experiment provides an introduction to human population genetics as participants learn to score genotypes and calculate genotype and allele frequencies. Students use safe saline mouthwash and Chelex® extraction to obtain their own DNA sample, amplify a 300-nucleotide Alu insertion into an intron of the H-cadherin gene, use electrophoresis to separate the 2 alleles, and determine their own genotype.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211367.pdf
This activity investigates whether the soy or corn ingredients in various processed foods contain a genetic modification. Students isolate DNA from wild-type and GM plant material (provided controls), and from food products of their choice. They use the extracted DNA as a template in 2 separate PCR reactions run under the same conditions
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211391.pdf
This kit explores an emerging trend in molecular biology and brings bioinformatics to the classroom in an accessible manner. Students take a new approach to taxonomy using “DNA barcodes”—short, unique DNA sequences—to learn about the biodiversity of plants, mammals, fish, or insects.
Bioinformatics Activities:
Bioinformatics Activity Updates:
Accessing the National Center for Biotechnology Information (NCBI) database, students use the Basic Local Alignment Search Tool (BLAST) to compare their sequences to gene segments published in the database. Using a case study approach, students continue to explore their sequences as they work to determine the location and role of their gene in a disease
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211391.pdf
Students go hands-on as they experience the power of silencing a single gene and learn how that method can be used to determine a gene’s function. This fascinating activity then culminates with a bioinformatics exercise.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211391.pdf
Students extract a sample of their own DNA and then amplify a 440-nucleotide segment of a hypervariable region of the mitochondrial chromosome, which contains numerous single nucleotide polymorphisms (SNPs). After gel electrophoresis confirms amplification, student samples may be sent for sequencing and students then compare their SNPs to ancient hominids and to people from different world populations.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211392.pdf
RNA interference (RNAi) is a technique that allows you to silence expression of a chosen gene by degrading the gene’s mRNA. This kit lets students use the Nobel Prize-winning technique to silence the dpy-13 gene in the non-parasitic roundworm C. elegans.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211233.pdf
This lab illustrates the use of DNA typing to identify individuals in court cases and disasters. It assays for variable numbers of tandem repeats (VNTR) polymorphisms, which are caused by short, repeated copies of a 16-nucleotide sequence at the pMCT118 locus. Differences in the number of repeated units produce longer and shorter alleles, which can be resolved by gel electrophoresis.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211403.pdf
Utilize methylation-sensitive enzymes to explore epigenetics—heritable changes in gene expression—that affect flowering in Arabidopsis. By growing wild-type Ler and mutant fwa-1 plants, students are able to observe phenotypic differences in flowering and then investigate this difference using multiple molecular techniques. Students conduct DNA extraction, restriction enzyme digest, PCR, gel electrophoresis, and bioinformatics to investigate the pivotal role DNA methylation plays in gene regulation.
Bioinformatics Activities:
Bioinformatics Activity Updates:
www.carolina.com/pdf/manuals/211377.pdf
This PTC Taster: Extraction, Amplification, and Electrophoresis Kit with CarolinaBLU® and 0.2-mL Tubes (with voucher) explores the molecular basis of the inherited ability to taste the bitter chemical phenylthiocarbamide (PTC). Students determine their ability to taste PTC using taste paper, use safe saline mouthwash and Chelex® extraction to obtain a sample of their own DNA, amplify a 221-nucleotide region of the PTC taste receptor gene, perform restriction digestion to differentiate the 2 alleles by gel electrophoresis, and determine how well the SNP genotypes actually correlate to tasting.

Which Genetic Organism Is Best for Your Classroom? With so many genetic organisms to choose from, it can be difficult to find the organism that

A History of DNA Barcoding DNA Barcodes: The beginning Paul Hebert of the University of Guelph coined the term DNA barcode to mean a unique

View the replay to learn practical classroom activities for students to build biotech skills. Even before the coronavirus global pandemic and the resulting need for
Get the latest news, free activities, teacher tips, product info, and more delivered to your inbox.